Gene expression is known to be a process that is regulated at multiple levels, and any dysfunctions can lead to aberrant cellular responses and deleterious defects. We focus on post-transcriptional mechanisms and question how regulation of mRNA translation or degradation can impact gene expression. We also question how aberrant/damaged mRNAs are degraded via ribosome-associated quality controls.
Using Saccharomyces cerevisiae as a model organism, we thus seek to explore new research fields through the characterization of factors, mechanisms and novel functions potentially conserved in plant and animal kingdoms.
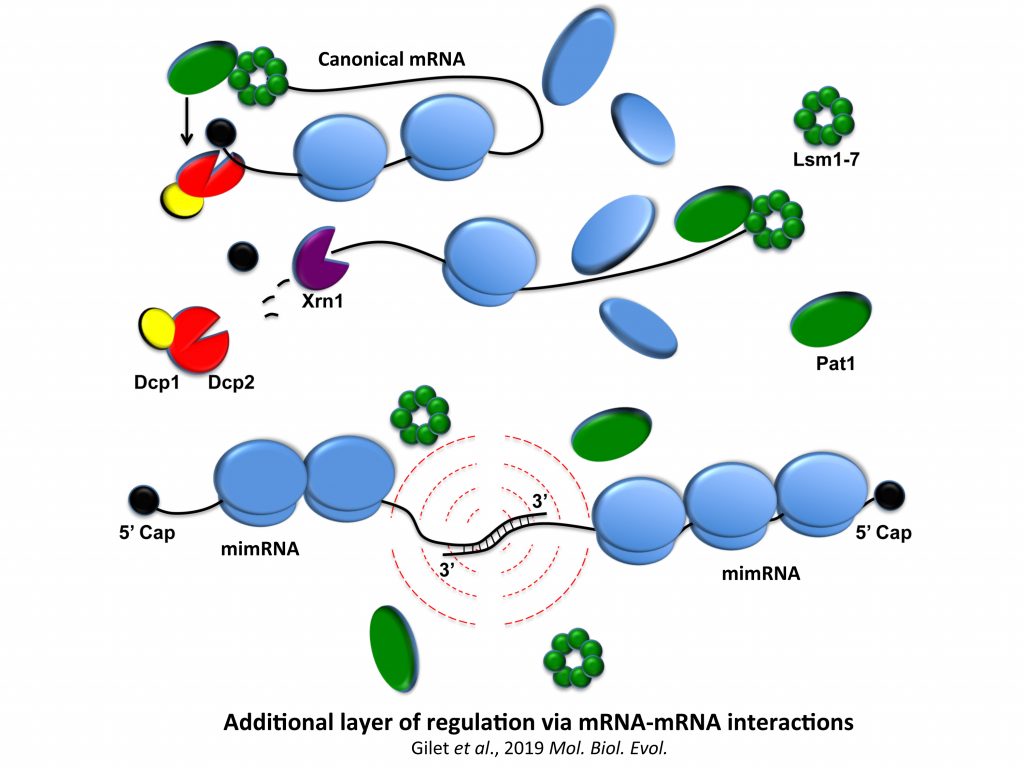
1/ A novel class of RNA-RNA interactions, mRNA-mRNA interactions.
We demonstrated the existence of a large class of RNAs, called mRNA-interacting mRNAs, mimRNAs, due to convergent and overlapping of gene transcriptions, potentially subjected to unusual mechanisms of post-transcriptional control in yeast, expected to be applicable to plant and human cells.
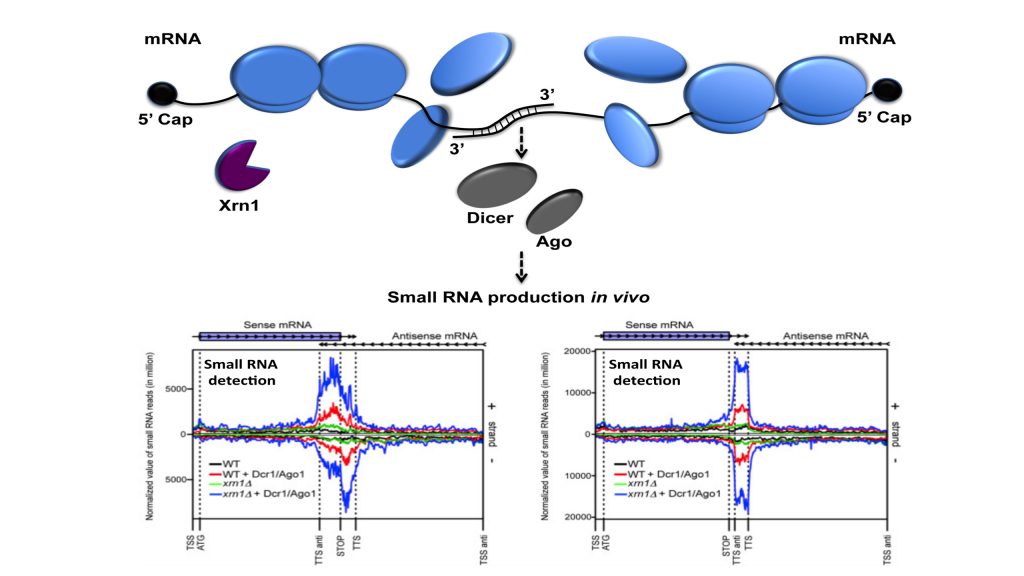
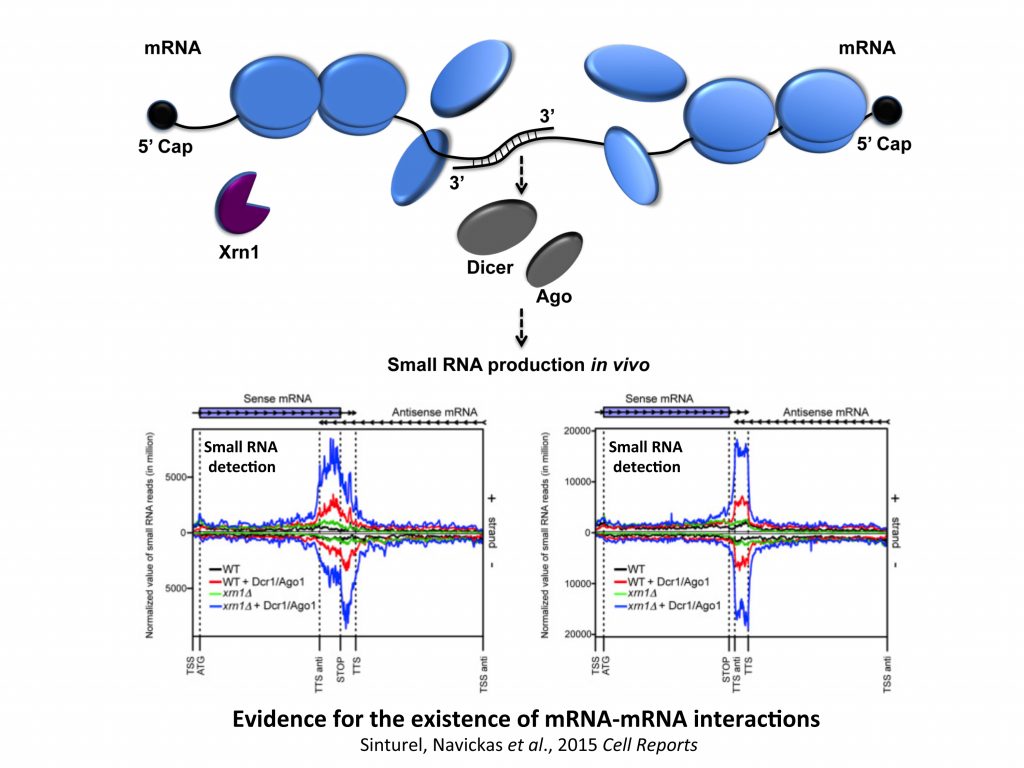
The starting point of this study was to understand why Xrn1 deficient cells have a peculiar post-transcriptional impact on the expression of the POR1 gene encoding a mitochondrial porin. This resulted in the demonstration that a defect in the activity of the 5’-3’ exoribonuclease Xrn1, the major 5’-3’ exoribonuclease highly conserved in eukaryotes, stabilizes convergent and 3’-overlapping transcripts that produce mRNA-mRNA interactions (potentially forming double-stranded RNAs, dsRNAs) detrimental for POR1 mRNA translation. This encouraged, in collaboration with A. Morillon’s group (Institut Curie), the genome-wide detection of all mRNA duplexes existing in xrn1 mutant and in wild-type cells. For this purpose, we expressed Dicer and Ago from S. castelli (a tool developed by David Bartel’s group, Drinnenberg et al., 2009 Science) to demonstrate the existence of mRNA duplexes (i.e. dsRNAs) in vivo and reveal the importance of Xrn1 in dsRNA formation and small RNA production in the presence of Dicer.
We now question the physiological importance of mRNA-mRNA interactions. We published a model of a post-transcriptional regulation mediated by mRNA duplex formation in which, upon stress, the translational repressor Pat1 binds preferentially to the 3′-UTR of canonical mRNAs (mRNAs not forming RNA-RNA interactions), limiting ribosome access on mRNA 5′-UTRs and promoting mRNA aggregation (i.e. promoting silencing) into granules. In contrast, messenger interacting mRNAs forming mRNA duplexes could escape Pat1 repression by masking 3’-UTR access and then fully participate in stress response. We also propose, in collaboration with I. Lafontaine (UMR7141), that these mRNA–mRNA interactions could confer an advantage along evolution.

2/ mRNA surveillance.
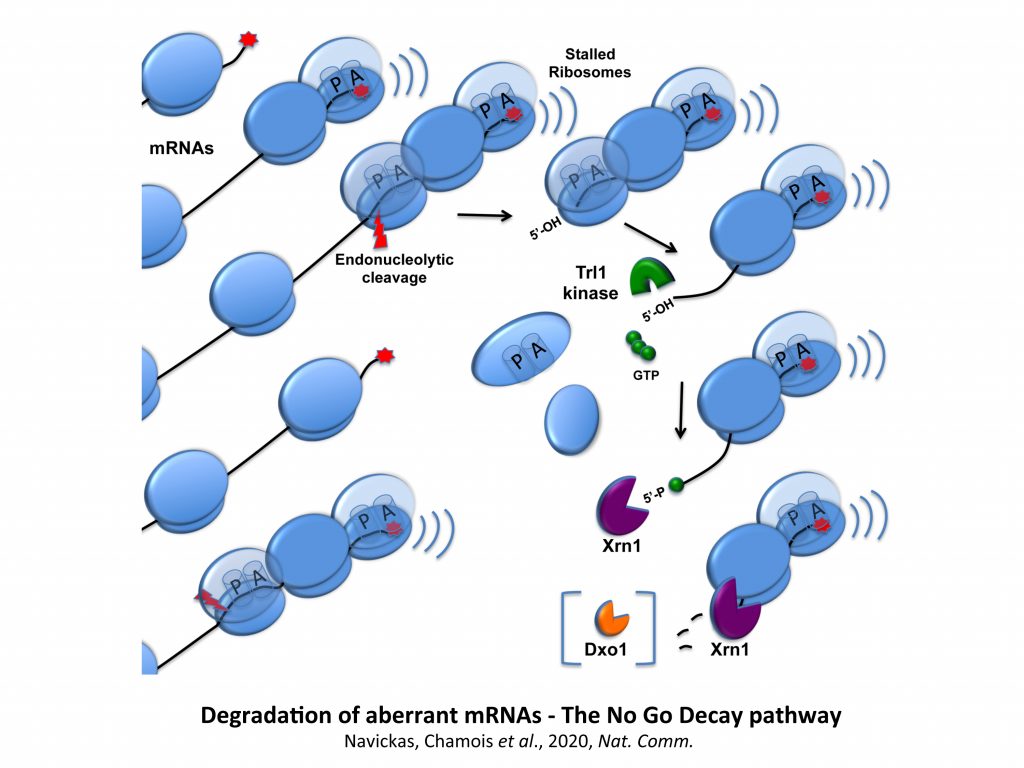
We have demonstrated that pairing between mRNAs occurs in S. cerevisiae and these mRNAs are potentially subjected to post-transcriptional regulations. Our exploratory studies suggested that, for a category of these messengers, mRNA-mRNA interactions mediate translational repression, with certain mRNA interactions being able to activate No-Go Decay (NGD) of mRNAs. NGD refers here to aberrant mRNA targeted for endonucleolytic cleavage and exonucleolytic digestion because of the stalling of elongating ribosomes.
All our observations above encouraged us to develop different approaches to better understand the mechanisms involved in NGD, being aware that the characterization of the factors cleaving these mRNAs has been a challenge for the last years. We thus have employed different genetic or biochemical approaches to identify specific factors acting in the vicinity of stalled ribosomes. We recently went further towards an understanding of the sequence of events that lead to the destruction of aberrant mRNAs by mapping the No Go endonucleolytic cleavage and by the demonstration of the importance of the Trl1 RNA kinase in the degradation of NGD RNA fragments.

3/ Regulation of ribonuclease activity.
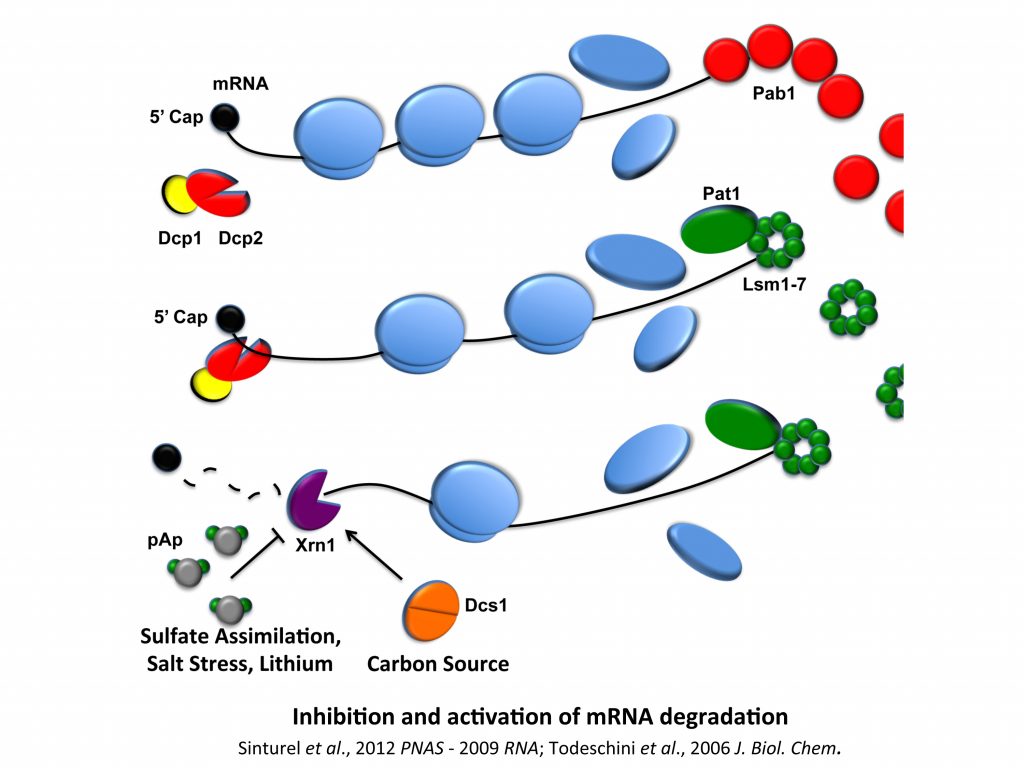
We have a long-term interest in analyzing the regulation of ribonuclease activity and in its importance in the modulation of gene expression when, for example, cells have to cope with environmental changes. We previously demonstrated that cells can limit Xrn1 inhibition by controlling pAp accumulation and that Dcs1 factor, whose expression is correlated to the nature of carbon sources, is a co-factor of Xrn1. We have set up fluorescent tools for the analysis of ribonuclease activities, and these projects have also been achieved by working within and in collaboration with C. Condon’s group (UMR8261).